
Introduction
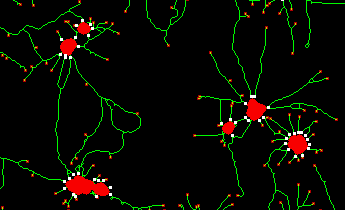
NeuriteQuant is a novel software tool for fully automated morphological analysis of large-scale image data from neuronal cultures. Please refer to the tutorials and documentation for detailed instructions, how to use NeuriteQuant. An example results database is provided to demonstrate analysis of drug induced changes in axons and dendrites of primary hippocampal neurons using NeuriteQuant. References are included to provide further examples and in-depth scientific discussion of the NeuriteQuant analysis toolkit and results obtained by its application.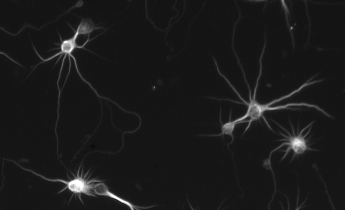
Features:
-
fully automated analysis of large data sets
- extraction of morphological parameters describing neurite outgrowth and branching
- generation of web-based data/image browser for easy screen review
- choice of interactive or manual analysis parameter determination
- freely available as open source
- Follow Tutorial 1 for a quick start into NeuriteQuant including installation and running morphological analysis on a test set.
- Follow Tutorials 2 and/or Tutorial 3 to learn how to adjust NeuriteQuant's analysis parameters.
- Read the documentation to learn about more detailed setting possibilities.
- Examine the NeuriteQuant script (NeuriteQuant_analysis.txt in the plugins/NeuriteQuant folder) to get basic information how to adjust/modify/extend NeuriteQuant for your specific needs.
- extraction of morphological parameters describing neurite outgrowth and branching
- generation of web-based data/image browser for easy screen review
- choice of interactive or manual analysis parameter determination
- freely available as open source
Getting Started:
- Check out the Example Databrowser from the Resources tab to examine if NeuriteQuant might suit your needs.- Follow Tutorial 1 for a quick start into NeuriteQuant including installation and running morphological analysis on a test set.
- Follow Tutorials 2 and/or Tutorial 3 to learn how to adjust NeuriteQuant's analysis parameters.
- Read the documentation to learn about more detailed setting possibilities.
- Examine the NeuriteQuant script (NeuriteQuant_analysis.txt in the plugins/NeuriteQuant folder) to get basic information how to adjust/modify/extend NeuriteQuant for your specific needs.
References
1. Dehmelt L., Poplawski G., Hwang E., Cho C., and Halpain S. NeuriteQuant: An Open Source Toolkit for High Content Screens of Neuronal Morphogenesis. submitted
Downloads |
| -NeuriteQuant1.23 (not compatible with ImageJ 1.43u) |
| recommended ImageJ(version
1.40g): |
| -for Windows |
| -forMacOS X |
| additional versions: |
| -NeuriteQuant1.24 (compatible with ImageJ 1.43u) |
Resources |
| -Tutorial1 |
| -Tutorial2 |
| -Tutorial3 |
| -Documentation |
| -Example Databrowser |
Links |
| -The Halpain Lab |
| -The Dehmelt Lab |
| -ImageJ home |